Examples
Examples of structure improvements are from the structure with Uniprot AC A4QBG9. The original structure from AlphaFold DB is shown in grey, and the structure optimised by FFFold is in colour.
Bond length
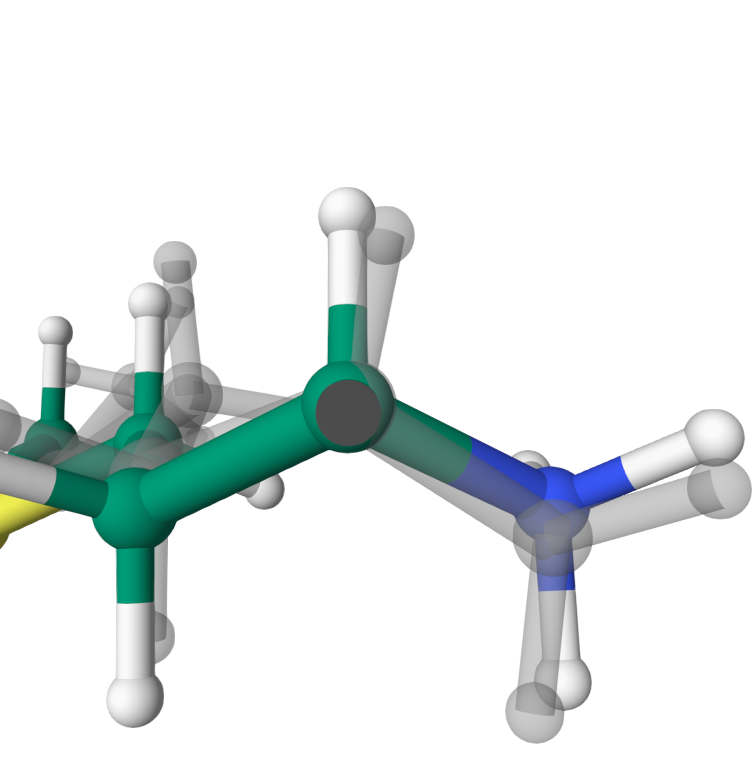
The bond length between CA and H atoms in MET1 is modified about 0.1 angstrom. On average, the bond lengths in MET1 are optimized by 0.05 angstrom.
Dihedral angles
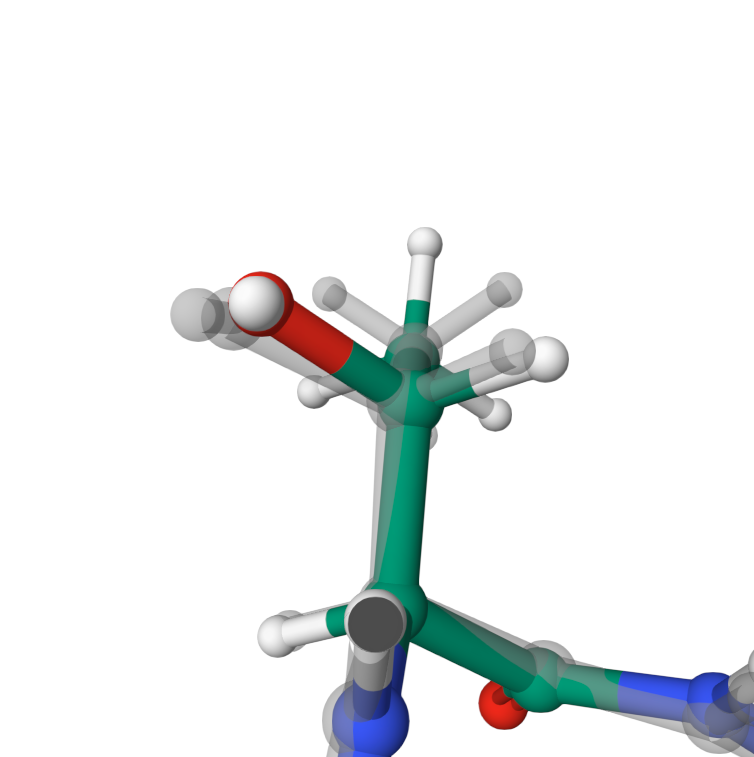
Dihedral angles in THR126 between atoms OG1, CB, CG2 and hydrogens HG21, HG22, HG23 are optimised from energetically unfavourable eclipsed
conformation.
Hydrogen bond
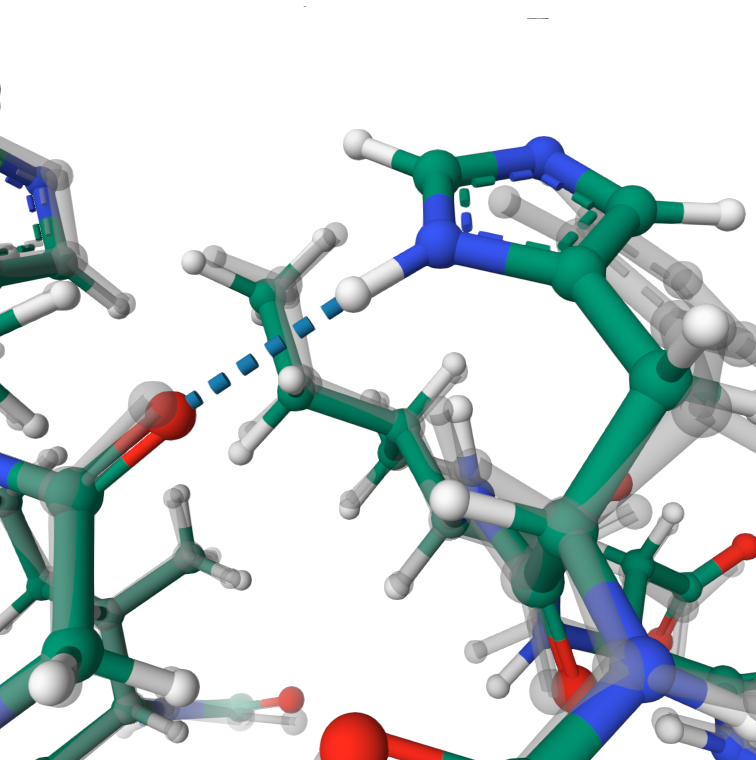
The hydrogen bond was formed between atoms HD1 from HIS20 and O from GLY86.
π–π stacking
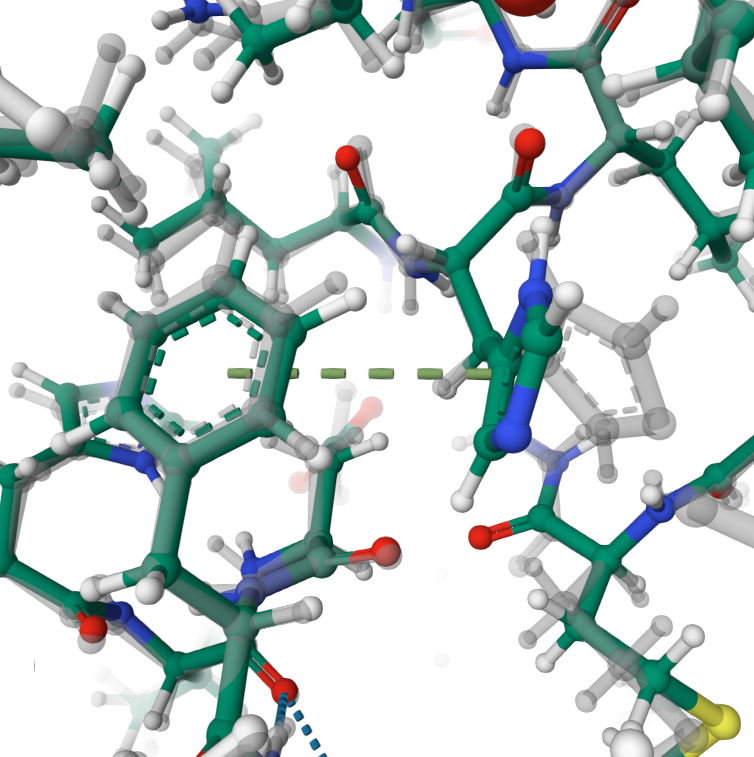
T-shaped π-π stacking is formed between residues PHE90 and HIS461.
FFFold tool is a part of services provided by ELIXIR – European research infrastructure for biological information.
For other services provided by ELIXIR's Czech Republic Node visit www.elixir-czech.cz/services.

Licence conditions in accordance with § 11 of Act No. 130/2002 Coll. The owner of the software is Masaryk
University, a public university, ID: 00216224. Masaryk University allows other companies and individuals to
use this software free of charge and without territorial restrictions in usual way, that does not depreciate
its value. This permission is granted for the duration of property rights. This software is not subject to
special information treatment according to Act No. 412/2005 Coll., as amended. In case that a person who
will use the software under this licence offer violates the licence terms, the permission to use the
software terminates.